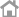TrioDx™ RT-PCR COVID-19 Test Kit
Clinomics TrioDx RT-PCR COVID-19 Test is an in vitro diagnostic real-time RT-PCR test intended for the
qualitative detection of nucleic acid from SARS-CoV-2 in nasopharyngeal, oropharyngeal, mid-turbinate, and
anterior nasal swabs collected from individuals suspected of COVID-19 by their healthcare provider.
The TrioDx is intended for use by qualified and trained clinical laboratory personnel specifically instructed in the
techniques of real-time PCR and in vitro diagnostic procedures. The TrioDx is only for use under the Food and
Drug Administration’s Emergency Use Authorization.
∙ Kit components(100 rxns/box)
The reagents included in one TrioDx kit are sufficient for testing 100 samples.
|
Name |
Volumes |
Quantity |
Storage upon receipt |
|
1
|
SARS-CoV-2 Positive Control
|
100 μL
|
1
|
- 20 ℃
|
|
2
|
2X Real-Time RT-PCR Mix
|
1,000 μL
|
1
|
- 20 ℃
|
|
3
|
10X Primer Probe Mix
|
200 μL
|
1
|
- 20 ℃
|
|
4
|
RNase Free H2O
|
500 μL
|
1
|
- 20 ℃
|
∙ Download

FDA-EUA

User Manual

Fact Sheet
(HCP&Patient)
Clinomics USA provides COVID-19 Testing using TrioDx kits in the Crestview Clinical Laboratory
Crestview Clinical Laboratory is a facility officially registered on the California COVID-19 Testing Task Force website.
Our laboratory capacity can processes hundreds of thousands of tests each year, and many thousands COVID-19
Testing per day for Public and Private Health service providers in Southern California.
Contact info : Justin A.Nguyen
(877) 429-6607
inguyen@crestviewlab.com
3 Target genes
RdRp gene +E gene +N gene
The TrioDx is a multiplex real-time reverse transcription polymerase chain reaction test (real-time RT-PCR). The SARS-CoV-2 Primer and Probe sets are designed
to detect RNA of the 2019-SARS nCoV in upper respiratory specimens from patients suspected of COVID-19 disease by their healthcare provider.
The nucleic acid sequences of primers and probes and their target genes used in the test are listed in the following table.
Table 1. Primer and probe sets and target genes
| Target |
Fluorescent dye |
Quencher |
|
RdRp
|
HEX
|
ZEN/IBFQ
|
|
E
|
TEXAS RED
|
ZEN/IBFQ
|
|
N
|
FAM
|
IBRQ
|
|
GAPDH (Internal Control)
|
Cy5
|
TAO/IBRQ
|
Analytical Accuracy
Limit of Detection (Analytical Sensitivity)
5 copies of viral RNA in a reaction (1 copy/ul in RNA extract[1]) for upper respiratory tract specimens
The LoD is defined as the lowest concentration of SARS-CoV-2 that can be reliably detected at least 95% of the time. Three LoD studies were performed to determine the
analytical sensitivity of the TrioDx: (1) a preliminary study with AccuPlex materials, (2) a confirmatory study with AccuPlex materials, and (3) a study with heatinactivated
virus (BEI Resources).
Final Reported LoD- summary of LoD for all three extraction methods The final reported LoD is summarized below in Table 18. The LoD based on the test algorithm is in the
bottom row of the table and shows the LoD is the same, 0.125 copies/μL, for all three extraction methods.
Table 18. Final reported LoD for each target for three different extraction methods
| Gene |
QIAamp Viral RNA Mini Kit (Manual Prep) |
Maxwell RSC Viral Total Nucleic Acid Kit with Maxwell RSC 48 instrument (Auto Prep) |
MagMAX Viral/Pathogen II Nucleic Acid Isolation Kit with KingFisher Flex instrument (Auto Prep) |
| RdRp gene |
0.25 copies/μL |
0.125 copies/μL |
0.25 copies/μL |
| E gene |
0.25 copies/μL |
0.125 copies/μL |
0.25 copies/μL |
| N gene |
0.25 copies/μL |
0.25 copies/μL |
0.25 copies/μL |
| Final reported LoD using all 3 genes |
0.125 copies/μL |
0.125 copies/μL |
0.125 copies/μL |
5 copies of viral RNA in a reaction (1 copy/ul in RNA extract[1]) for upper respiratory tract specimens
The LoD is defined as the lowest concentration of SARS-CoV-2 that can be reliably detected at least 95% of the time. Three LoD studies were performed to determine the
analytical sensitivity of the TrioDx: (1) a preliminary study with AccuPlex materials, (2) a confirmatory study with AccuPlex materials, and (3) a study with heatinactivated virus (BEI Resources).
Final Reported LoD- summary of LoD for all three extraction methods The final reported LoD is summarized below in Table 18. The LoD based on the test algorithm is in the
bottom row of the table and shows the LoD is the same, 0.125 copies/μL, for all three extraction methods.
Table 18. Final reported LoD for each target for three different extraction methods
| Gene |
QIAamp Viral RNA Mini Kit (Manual Prep) |
Maxwell RSC Viral Total Nucleic Acid Kit with Maxwell RSC 48 instrument (Auto Prep) |
MagMAX Viral/Pathogen II Nucleic Acid Isolation Kit with KingFisher Flex instrument (Auto Prep) |
| RdRp gene |
0.25 copies/μL |
0.125 copies/μL |
0.25 copies/μL |
| E gene |
0.25 copies/μL |
0.125 copies/μL |
0.25 copies/μL |
| N gene |
0.25 copies/μL |
0.25 copies/μL |
0.25 copies/μL |
| Final reported LoD using all 3 genes |
0.125 copies/μL |
0.125 copies/μL |
0.125 copies/μL |
The TrioDx has not been FDA cleared or approved; but has been authorized by FDA under an EUA for use in authorized laboratories.
Testing authorized by the U.S. FDA is limited to laboratories that are certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, and
that meet requirements to perform high complexity tests.
Inclusivity for new variants
Additional inclusivity of the TrioDx was evaluated in silico by comparing the primer/probe sequences separately with each of 9,275 new variant SARS-CoV-2 sequences
published in GISAID databases (through January 25th, 2021).
Three representative Emerging Variants lineages were used from:
https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html.
This included 31 cases in P.1 (Brazil), 8,573 cases in B.1.1.7 (United Kingdom), and 671 cases in B.1.351 (South Africa). Each new variant SARS-CoV-2 sequence was separately
used for alignment with the TrioDx assay.
The SARS-CoV-2 TrioDx primers and probes for RdRp, E and N gene targets showed 100% homology with reference sequence (NC_045512.2). The inclusivity is summarized
in Table 20.
Table 20. Inclusivity test results for new variants: P.1 (Brazil), B.1.1.7 (United Kingdom), B.1.351 (South Africa)
| Database |
Target Genes |
Homology |
| Forward Primer |
Number of sequences in databases without 100% homology |
Probe |
Number of sequences in databases without 100% homology |
Reverse Primer |
Number of sequences in databases without 100% homology |
| GISAID (n=9275) |
RdRp gene |
99.99 % |
1 |
99.81 % |
18 |
99.96 % |
4 |
| E gene |
99.36 % |
59 |
99.92 % |
7 |
100 % |
0 |
| N gene |
99.62 % |
35 |
98.71 %(CASE 3) |
120 |
99.83 % |
16 |
Additional inclusivity of the TrioDx was evaluated in silico by comparing the primer/probe sequences separately with each of 9,275 new variant SARS-CoV-2 sequences
published in GISAID databases (through January 25th, 2021).
Three representative Emerging Variants lineages were used from:
https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html.
This included 31 cases in P.1 (Brazil), 8,573 cases in B.1.1.7 (United Kingdom), and 671 cases in B.1.351 (South Africa). Each new variant SARS-CoV-2 sequence was separately
used for alignment with the TrioDx assay.
The SARS-CoV-2 TrioDx primers and probes for RdRp, E and N gene targets showed 100% homology with reference sequence (NC_045512.2). The inclusivity is summarized
in Table 20.
Table 20. Inclusivity test results for new variants: P.1 (Brazil), B.1.1.7 (United Kingdom), B.1.351 (South Africa)
| Database |
Target Genes |
Homology |
| Forward Primer |
Number of sequences in databases without 100% homology |
Probe |
Number of sequences in databases without 100% homology |
Reverse Primer |
Number of sequences in databases without 100% homology |
| GISAID (n=9275) |
RdRp gene |
99.99 % |
1 |
99.81 % |
18 |
99.96 % |
4 |
| E gene |
99.36 % |
59 |
99.92 % |
7 |
100 % |
0 |
| N gene |
99.62 % |
35 |
98.71 %(CASE 3) |
120 |
99.83 % |
16 |
Clinical Evaluation (Clinical Sensitivity)
100 % of clinical accuracy/sensitivity from 200,000+ Clinical samples
100 % of clinical accuracy/sensitivity from
200,000+ Clinical samples
The clinical performance of the TrioDx was established by testing retrospective samples using 30 natural, noncontrived positive clinical samples and 30 natural, non-contrived
negative clinical samples that were procured from a reference laboratory. Commercially sourced positive clinical samples were used following the FDA's COVID-19 Test
Development and Review FAQs. All 60 clinical specimen samples were Nasopharyngeal Swabs (NP) in Aptima Specimen Transport Medium or Multitrans Medium.
All procedures for the TrioDx clinical evaluation were conducted in accordance with Section 10 (Test Procedures) described in this Instruction for Use (IFU).
The results of the clinical evaluation study are summarized in Table 23. Both positive percent agreement (PPA) and negative percent agreement (NPA) came out to be 100%
Table 22. Clinical evaluation results summary
| n = 67 |
EUA Authorized Comparator Test |
Total |
| Positive |
Negative |
| TrioDx |
Positive |
37 |
0 |
37 |
| Negative |
0 |
30 |
30 |
| Total |
37 |
30 |
67 |
Positive Percent Agreement: 100% (37/37) 95% CI: 90.51% - 100.00%
Negative Percent Agreement: 100% (30/30) 95% CI: 88.43% - 100.00%
Test Procedure


Contact Information
Sumin Choi ┃ Sales & Marketing Director
+1 (917) 617-9481 smchoi@clinomics.com
Clinomics USA Inc.
8949 Complex Drive B, San Diego, CA 92123